SHARED GENETIC FACTORS IN HELICOBACTER PYLORI-INDUCED GASTRITIS AND GASTRIC CANCER: A NETWORK-DRIVEN ANALYSIS OF GENE EXPRESSION DATA
DOI:
https://doi.org/10.71146/kjmr356Abstract
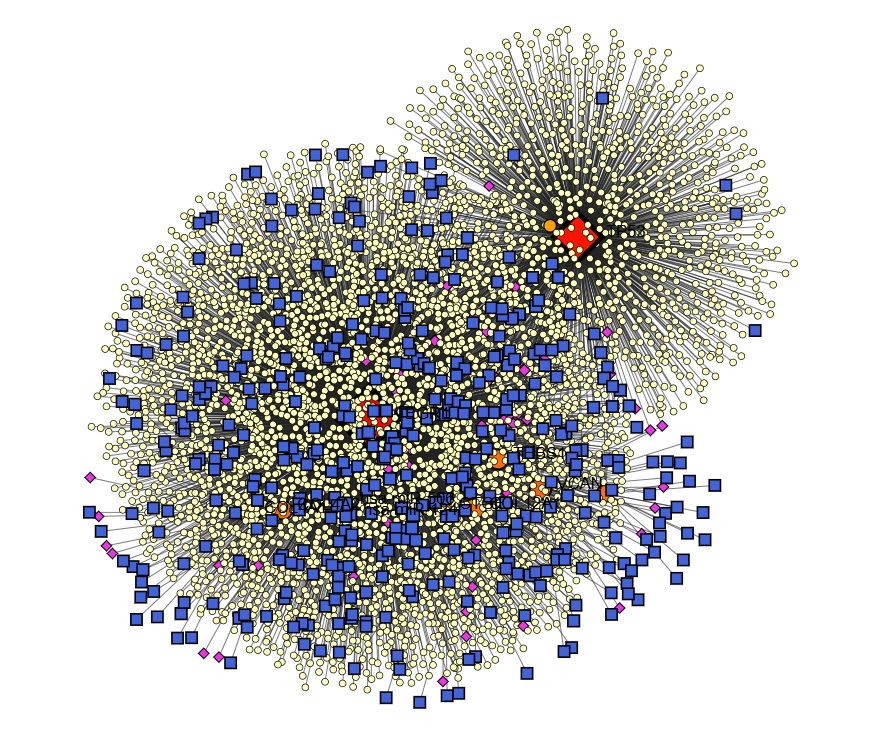
Two of the most common medical conditions in the world, gastritis and stomach cancer, are caused by the Helicobacter pylori (H. pylori). Gastric cancer and H. pylori-induced gastritis has both been the subject of molecular research, but the latter has received much more interest. In attempt to identify shared genetic factors that contribute to various diseases, this study examines gene expression data from the public microarray data set known as the Gene Expression Omnibus (GEO) database. Three different types of stomach cancer were investigated: GSE54129, GSE65168 and GSE13911. Using GEO2R, we identified genes that were overexpressed in both diseases. In order to get a better understanding of how genes work, we conducted GO and pathway enrichment investigations. In order to identify hub genes and important molecular interactions, a STRING-based PPI network was constructed using Cytoscape. To investigate hub genes, we used CytoHubba and MCODE to look at important chemical complexes and strongly connected nodes. The investigation of gene regulation was conducted using Network Analyst, which allowed us to construct TF-gene and TF-miRNA regulatory networks. Curing gastritis and stomach cancer caused by H. pylori might be possible with the use of a Drug Bank medication interaction network. We may be able to better understand and treat stomach issues caused by H. pylori if we use an integrated bioinformatics approach. It has the ability to identify genetic circuits, biomarkers and therapeutic targets.
Downloads
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Abuzar Mehdi Khan, Ayesha Iram, Muhammad Adnan, Muhammad Imran Ul Haq, Waseem Abbas, Muhammad Imran Malik (Author)

This work is licensed under a Creative Commons Attribution 4.0 International License.






